Yeasen
Hieff NGS DNA Selection Beads
Hieff NGS DNA Selection Beads
Couldn't load pickup availability
Description
Hieff NGS™ DNA Selection Beads are prepared based on the SPRI (Solid Phase Reverse Immobilization) principle and is applicable for DNA purification and size selection during the preparation of next generation sequencing (NGS) libraries. Hieff NGS™ DNA Selection Beads is compatible with various of DNA and RNA library prep kits and is a good competitor of AMPure beads.
Components
| Components No. | Name | 12601ES08 | 12601ES56 | 12601ES75 |
| 12601 | Hieff NGS™ DNA Selection Beads | 5 mL | 60 mL | 450 mL |
Specifications
| Product Line | DNA clean and selection Beads |
| Starting Material | DNA |
| Compatibility | DNA |
| Isolation Technology | Magnetic Bead |
| Final Product Type | DNA |
| For Use With (Application) | DNA celan up, DNA size slection |
Shipping and Storage
The beads are shipping with ice packs and can be stored at 2°C-8°C for one year.
Instructions
- 1. Preparation
Equilibrate the selection beads at room temperature for at least 30 min before use.
- 2. Size selection
The operation flow of size selection is shown in Figure 1 and the protocol is as follows.

Figure 1. The Operation Flowchart of Size Selection
2.1Mix the beads thoroughly by vortexing or pipetting up and down every time before using.
2.2Add the first round of selection beads to the sample (refer to Table 1). Mix thoroughly by vortexing or pipetting up and down at least 10 times.
2.3Incubate at room temperature for 5 min.
2.4Spin down the tube briefly and place it on magnetic stand. When the solution is clear (about 5 min), transfer the supernatant to a new PCR tube.
2.5Add the second round of selection beads to the sample from step 2.4 according to Table 1. Mix thoroughly by vortexing or pipetting up and down at least 10 times.
2.6Incubate at room temperature for 5 min.
2.7Spin down the tube briefly and place it on magnetic stand. When the solution is clear (about 5 min), aspirate the supernatant and discard.
2.8Keep the tube in the magnetic stand and add 200 μL of freshly prepared 80% ethanol to without disturbing the beads, incubate at room temperature for 30 sec. Aspirate the ethanol and discard.
2.9Repeat step 2.8 once for a total of two washes.
2.10Remove residual ethanol with 10 µL pipette tips. Keep the tube in the magnetic stand, air dry the selection beads with the lid open until cracks just appear (about 5 min).
Note:Donotover-drytheselectionbeads.ThismayresultinlowerrecoveryDNAtarget.
2.11Remove the tube from the magnetic stand. Add an appropriate amount of ddH2O (≥20 µL) and mix thoroughly by vortexing or pipetting up and down at least 10 times.
2.12Incubate at room temperature for 5 min.
Spin down the tube briefly and place it on the magnetic stand. When the solution is clear (about 5 minutes), transfer 20 μL of the supernatant to a new tube.
- 3.Recommended Conditions for DNA Size Selection
The calf thymus DNA was fragmented by sonication to prepare a fragment of 100-1,000 bp, and two rounds of size selection were performed according to Table 1. The results were analyzed using Agilent 2100 Bioanalyzer (Figure 2).
Table 1. Recommended condition for DNA size selection
| Length of DNA fragment | 250-350 bp | 320-420 bp | 450-550 bp | 550-700 bp | 700-900 bp | 800-1,000 bp |
| Ratio of Beads: DNA for the 1st Round | 0.80× | 0.70× | 0.60× | 0.55× | 0.50× | 0.45× |
| Ratio of Beads: DNA for the 2nd Round | 0.20× | 0.20× | 0.20× | 0.15× | 0.15× | 0.15× |
Note: "×" in the table indicates the volume of sample DNA. For example, if the insert length of the library is 250 bp and the sample DNA volume is 100 μL, the volume of magnetic beads used in the first round of sorting is 0.80×100 μL=80 μL; the volume of magnetic beads used in the second round of sorting is 0.20× 100 μL=20 μL.
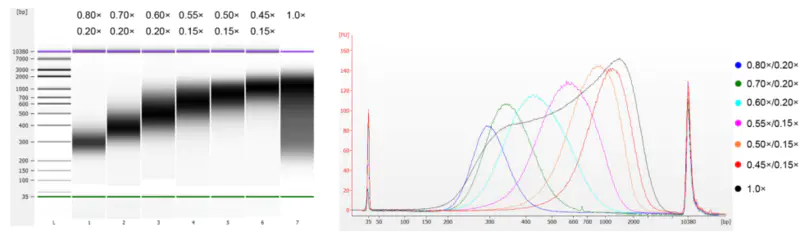
Figure 2. Agilent 2100 high-sensitivity DNA chip electropherogram
Notes:
1. For your safety and health, please wear lab coats and disposable gloves for operation.
Documents:
